A few days ago I wrote about how to solve differential equations with SciPy’s ivp_solve function using Van der Pol’s equation as the example. Van der Pol’s equation is
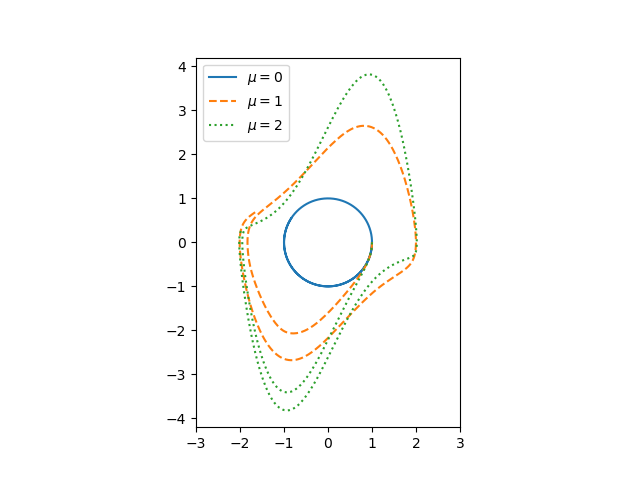
The parameter μ controls the amount of nonlinear damping. For any initial condition, the solution approach a periodic solution. The limiting periodic function does not depend on the initial condition [1] but does depend on μ. Here are the plots for μ = 0, 1, and 2 from the previous post.
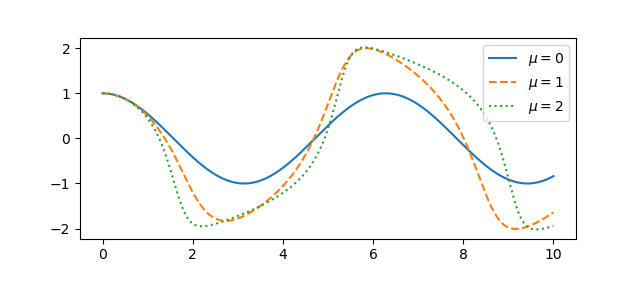
A couple questions come to mind. First, how quickly do the solutions become periodic? Second, how does the period depend on μ? To address these questions, we’ll use an optional argument to ivp_solve we didn’t need in the earlier post.
Using events in ivp_solve
For ivp_solve an event is a function of the time t and the solution y whose roots the solver will report. To determine the period, we’ll look at where the solution is zero; our event function is trivial since we want to find the roots of the solution itself.
Recall from the earlier post that we cast our second order ODE as a pair of first order ODEs, and so our solution is a vector, the function x and its derivative. So to find roots of the solution, we look at what the solver sees as the first component of the solver. So here’s our event function:
def root(t, y): return y[0]
Let’s set μ = 2 and find the zeros of the solution over the interval [0, 40], starting from the initial condition x(0) = 1, x‘(0) = 0.
mu = 2
sol = solve_ivp(vdp, [0, 40], [1, 0], events=root)
zeros = sol.t_events[0]
Here we reuse the vdp function from the previous post about the Van der Pol oscillator.
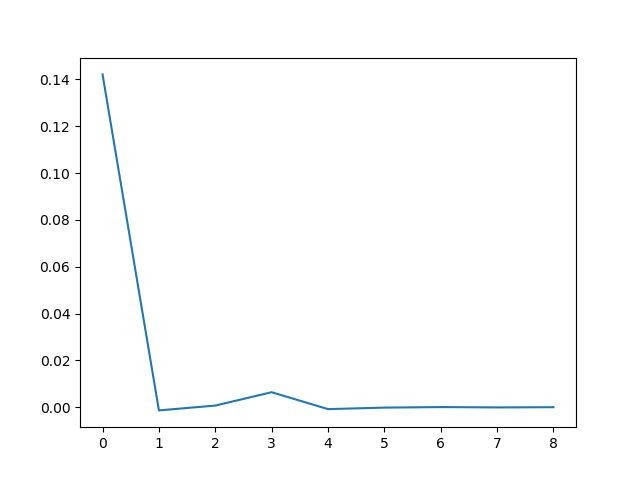
To estimate the period of the limit cycle we look at the spacing between zeros, and how that spacing is changing.
spacing = zeros[1:] - zeros[:-1]
deltas = spacing[1:] - spacing[:-1]
If we plot the deltas we see that the zero spacings quickly approach a constant value. Zero crossings are half periods, so the period of the limit cycle is twice the limiting spacing between zeros.
Theoretical results
If μ = 0 the Van der Pol oscillator reduces to a simple harmonic oscillator and the period is 2π. As μ increases, the period increases.
For relatively small μ we can calculate the period as above, but as μ increases this becomes more difficult numerically [2]. But one can easily show that the period is asymptotically
T ~ (3 – 2 log 2) μ
as μ goes to infinity. A more refined estimate due to Mary Cartwright is
T ~ (3 – 2 log 2) μ + 2π/μ1/3
for large μ.
More VdP posts
[1] There is a trivial solution, x = 0, corresponding to the initial conditions x(0) = x‘(0) = 0. Otherwise, every set of initial conditions leads to a solution that converges to the periodic attractor.
[2] To see why large values of μ are a problem numerically, here’s a plot of a solution for μ = 100.
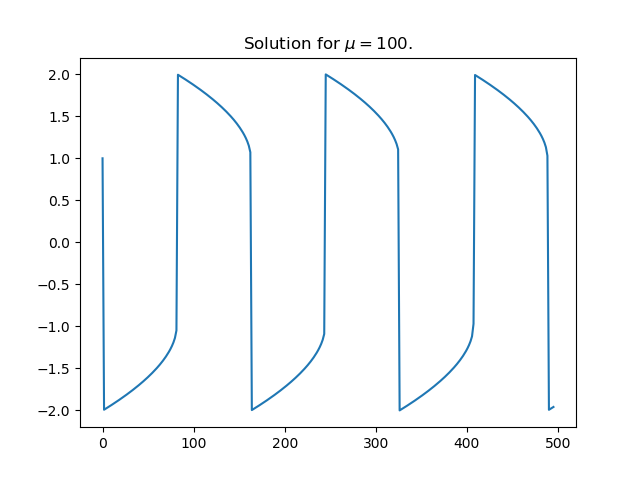
The solution is differentiable everywhere, but the derivative changes so abruptly at the maxima and minima that it is discontinuous for practical purposes.