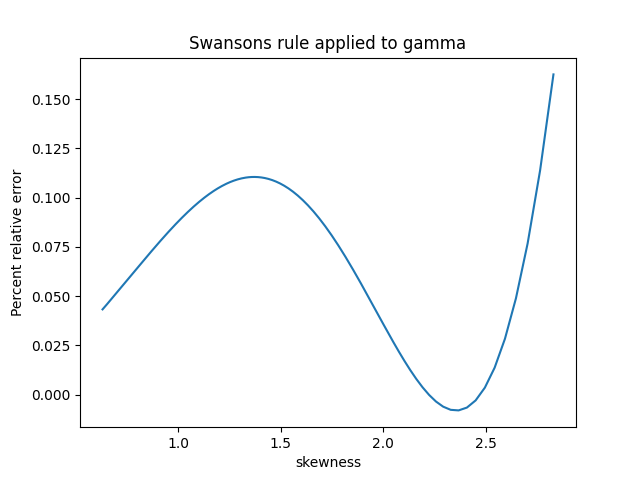
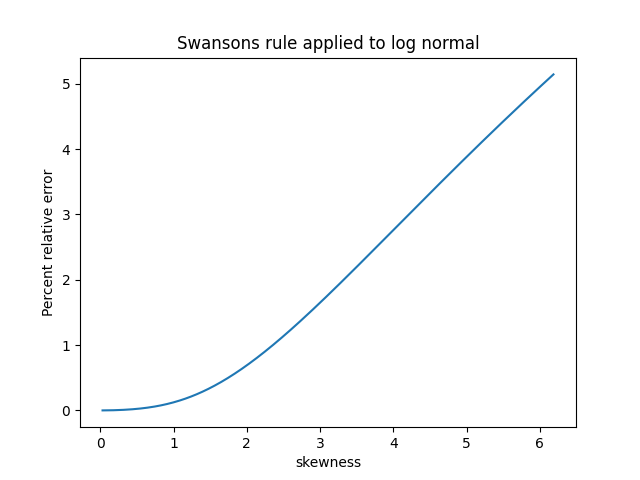
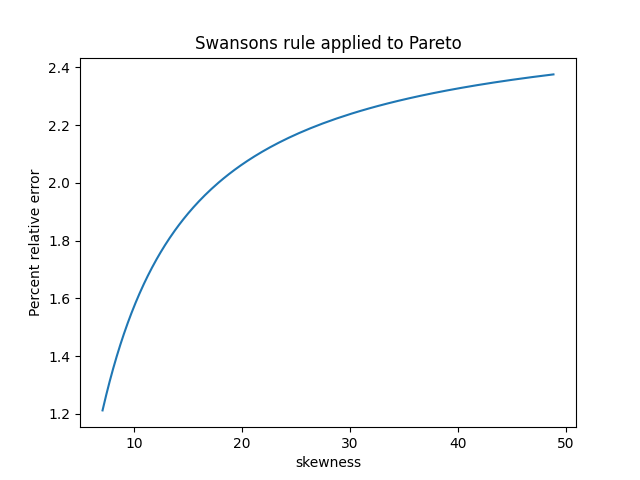
The previous post showed that the standard deviation of a sample of size n can be well estimated by multiplying the sample range by a constant dn that depends on n.
The method works well for relatively small n. This should sound strange: typically statistical methods work better for large samples, not small samples. And indeed this method would work better for larger samples. However, we’re not so much interested in the efficiency of the method per se, but its efficiency relative to the standard way of estimating standard deviation. For small samples, both methods are not very accurate, and the two methods appear to work equally well.
If we want to use the range method for larger n, there are a couple questions: how well does the method work, and how do we calculate dn.
Simple extension
Ashley Kanter left a comment on the previous post saying
dn = 3 log n0.75 (where the log is base 10) seems to perform quite well even for larger n.
Ashley doesn’t say where this came from. Maybe it’s an empirical fit.
The constant dn is the expected value of the range of n samples from a standard normal random variable. You could find a (complicated) expression for this and then find a simpler expression using an asymptotic approximation. Maybe I’ll try that later, but for now I need to wrap up this post and move on to client work.
Note that
log10 n0.75 = 0.977 loge(n)
and so we could use
dn = log n
where log is natural log. This seems like something that might fall out of an asymptotic approximation. Maybe Ashley empirically discovered the first term of a series approximation.
Update: See this post for a more detailed exploration of how well log n, square root of n, and another method approximate dn.
Update 2: Ashley Kanter’s approximation was supposed to be 3 (log10 n) 0.75 rather than 3 log10 (n0.75) and is a very good approximation. This is also addressed in the link in the first update.
Simulation
Here’s some Python code to try things out.
import numpy as np
from scipy.stats import norm
np.random.seed(20220309)
n = 20
for _ in range(5):
x = norm.rvs(size=n)
w = x.max() - x.min()
print(x.std(ddof=1), w/np.log(n))
And here are the results.
| std | w/d_n |
|-------+-------|
| 0.930 | 1.340 |
| 0.919 | 1.104 |
| 0.999 | 1.270 |
| 0.735 | 0.963 |
| 0.956 | 1.175 |
It seems the range method has more variance, though notice in the fourth row that the standard estimate can occasionally wander pretty far from the theoretical value of 1 as well.
We get similar results when we increase n to 50.
| std | w/d_n |
|-------+-------|
| 0.926 | 1.077 |
| 0.889 | 1.001 |
| 0.982 | 1.276 |
| 1.038 | 1.340 |
| 1.025 | 1.209 |
Not-so-simple extension
There are ways to improve the range method, if by “improve” you mean make it more accurate. One way is to divide the sample into random partitions, apply the method to each partition, and average the results. If you’re going to do this, partitions of size 8 are optimal [1]. However, the main benefit of the range method [2] is its simplicity.
Related posts
[1] F. E. Grubbs and C. L. Weaver (1947). The best unbiased estimate of a population standard deviation based on group ranges. Journal of the American Statistical Association 42, pp 224–41
[2] The main advantage now is its simplicity, When it was discovered, the method reduced manual calculation, and so it could have been worthwhile to make the method a little more complicated as long as the calculation effort was still less than that of the standard method.